
In the realm of genetic research, DNA FISH has long been a stalwart technique for unraveling chromosomal mysteries, shedding light on chromatin organization, and diagnosing clinical abnormalities. However, its reliance on heat denaturation and the need for lengthy target regions have posed significant obstacles, particularly when dissecting fine chromatin structures below 10 kilobases (kb), such as those introduced by exogenous genes during cell and gene therapy.
CRISPR-Cas9, a formidable tool derived from Streptococcus pyogenes, extensively explored for scrutinizing genome structures both in vivo and in vitro. The CRISPR-Cas9-sgRNA ribonucleoprotein complex, known for its precision in targeting specific genome loci, presented an opportunity to revolutionize the conventional DNA FISH methodology.
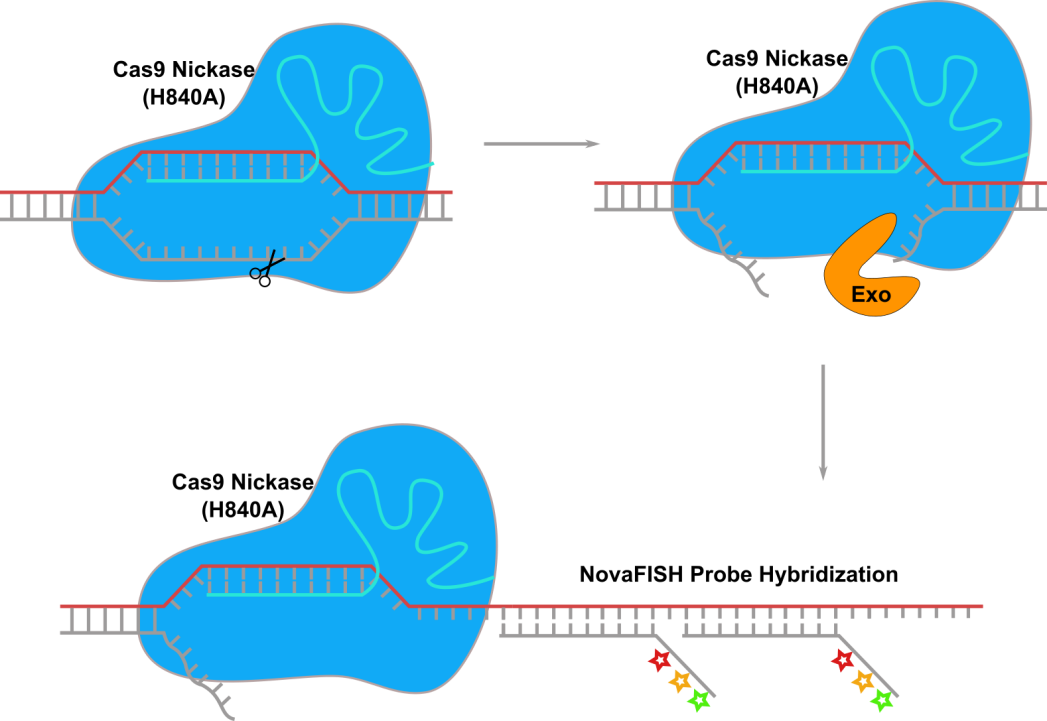
Figure 1. CRISPR-ExoFISH workflow, Genomic DNA loci is binded by Cas9 nickase(H840A)-sgRNA (cyan) ribonucleoprotein complex and cut at NTS strand (grey). Exonuclease find the flapped DNA on NTS strand and digest the NTS stand. Then NovaFISH probe is hybridizing to the TS stand (red).
We introduces a novel approach, termed CRISPR-ExoFISH—a pioneering approach designed to open genome loci and digest specific regions into single-strand DNA (ssDNA), ensuring perpetual accessibility for DNA FISH probes. Unlike its traditional counterparts, CRISPR-ExoFISH eliminates the need for heat denaturation, requiring only a compact region below 5 kb for DNA FISH probes to bind and generate specific signals.
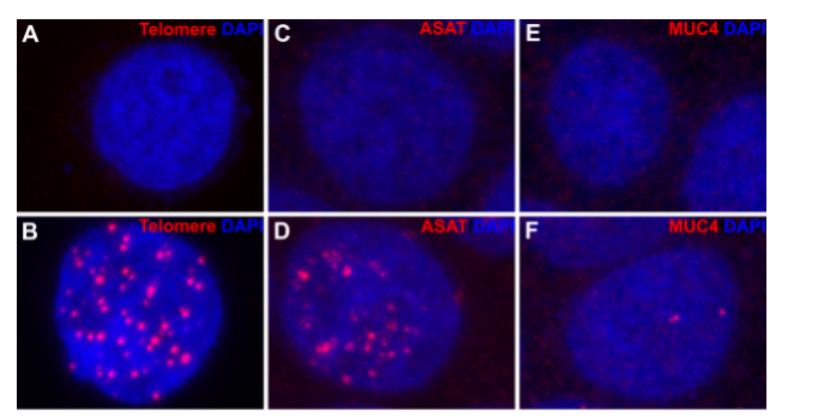
Fiqure 2. CRISPR-ExoFISH of Human genomic targets. (A, C, E) Hela cell stained with NovaFISH probe for telomere, ASAT, and MUC4 respectively. (B, D, F) Hela cell were fixed and stained with NovaFlSH probe against Human telomere, ASAT, and MUC4 respectively, after genomic DNA of telomere, ASAT and MUC4 is opened and digested as ssDNA.
In a groundbreaking test on human cells, CRISPR-ExoFISH showcased its prowess on three distinct targets. Telomere analysis (Figure 2A-B) revealed the efficacy of telomere sgRNA in facilitating efficient hybridization without the reliance on heat denaturation. The ssDNA nature of telomeres post-Cas9-sgRNA binding and T7 exonuclease digestion became apparent, marking a significant advancement.
Further validation came with the examination of the alpha satellite DNA (ASAT) repeat region (Figure 2C-D), displaying specific staining in all repeated genomic regions, underscoring the method's versatility. CRISPR-ExoFISH transcended its application on repeat regions, successfully detecting non-repetitive genomic loci, as demonstrated in the validation of MUC4 intron 1 (Figure 2E-F). The specific staining of the MUC4 gene intron 1 showcased the method's adaptability and potential for diverse genomic analyses.
In conclusion, CRISPR-ExoFISH emerges as a transformative force in the genetic research landscape, offering a streamlined and efficient alternative to traditional DNA FISH methods. Its ability to unlock the mysteries of fine chromatin structures without the need for heat denaturation opens new avenues for exploring genomic intricacies and holds great promise for advancing diagnostic and therapeutic applications.

