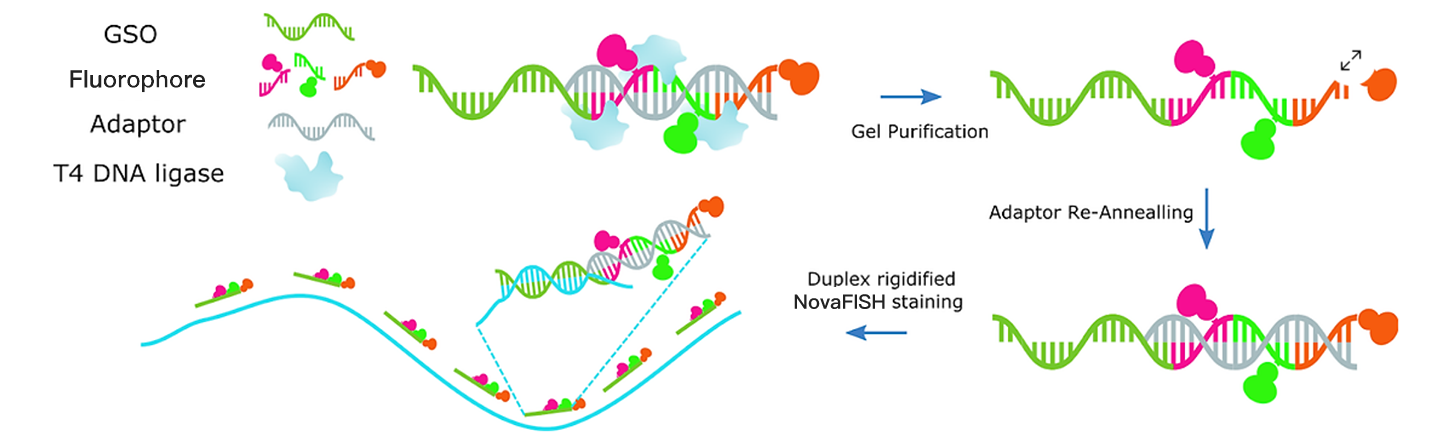
NovaFISH is a new kind of smFISH (single molecule FISH) technique. It is the industry’s first enzymatic labeled smFISH probe. After the selection of various enzymes which could link DNA molecules together, NovaFISH ends up having a T4 DNA ligase to conjugate multiple prelabeled fluorescent oligos (Nova) onto gene-specific oligo (GSO). The multiple labeled NovaFISH probes can detect a large number of gene expression at one hybridization round. The multiplexity follows an exponential growth with the number of fluorophores available (i.e. the number of channels in the microscope).
For your application, you need to prepare the cell or tissue sample by fixation with formaldehyde. After membrane permeabilization with ethanol or Triton X-100, the NovaFISH probe can be readily diffused into the cell and bind with the specific RNA target. On average, our NovaFISH probe can bind to the RNA with a probability higher than 90% because its design algorithm optimally selects shorter DNA sequences with higher binding capacity. As a result, your smFISH dot under the microscope is brighter due to the higher number of fluorophores concentrated on the RNA target.
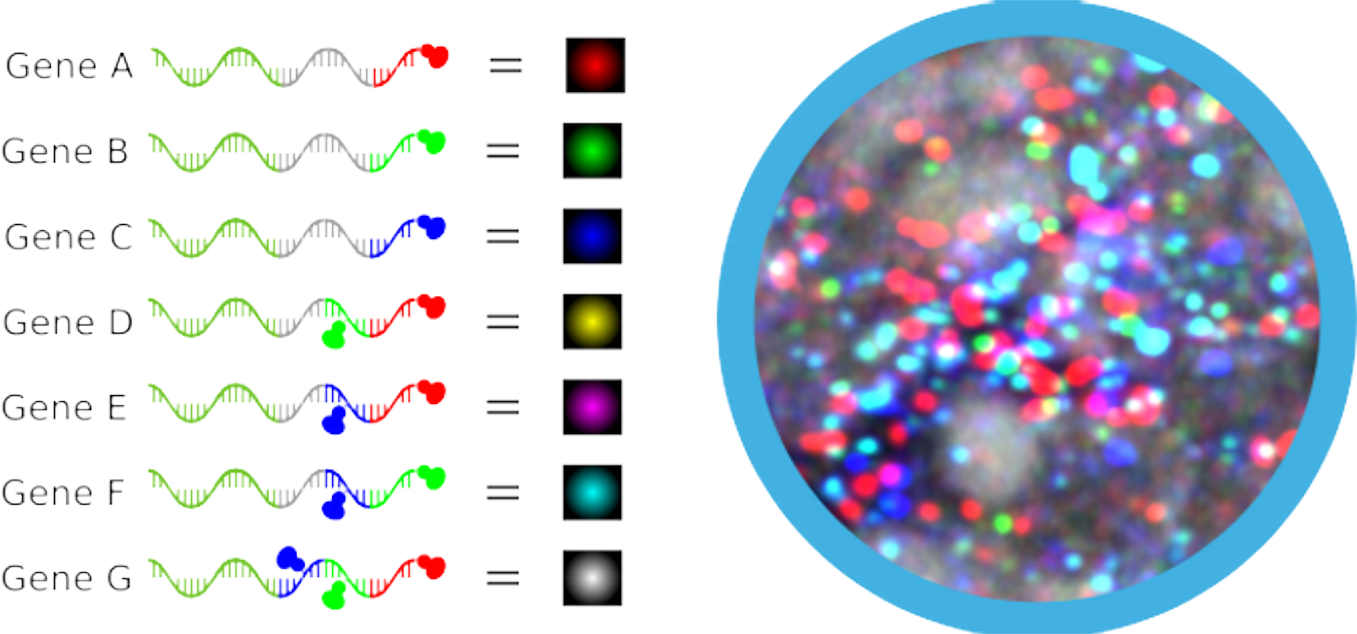
We have successfully validated NovaFISH on a Zeiss confocal microscope (LSM800 with Airyscan) with three lasers to detect seven genes simultaneously. With the full combination of three base colors, it is now possible to detect seven genes in one hybridization reaction and the resulting multi-channel imaging data can be decoded by our AI-powered quantitation software NovaREAD.
In principle, our proprietary NovaFISH can provide a precise gene expression profiling at a single molecule level up to 127 targets with a state-of-art microscope with 7 lasers. To further push this limit, it is also compatible with sequential FISH if more targets need to be validated.
If you would like to know more about our NovaFISH technology, please write to us at order@pixelbiosciences.com

