
In October 2019, Andrew Anzalone, a postdoctoral fellow in David Liu's lab, and colleagues unveiled a groundbreaking innovation in genetic engineering known as Prime Editing, in a publication featured in Nature. This development represents a significant stride forward in the realm of genetic manipulation, offering a highly precise and versatile method that addresses several limitations encountered with conventional CRISPR-Cas9 systems. Let's delve into the essence of Prime Editing, exploring its functionality and several advantages.
Prime Editing introduces a novel approach to genetic modification, characterized as a "search-and-replace" technique that facilitates targeted insertions, deletions, and all conceivable base-to-base conversions. Unlike traditional CRISPR-Cas9 methods, Prime Editing achieves these alterations without the necessity for double-strand breaks (DSBs) or reliance on donor DNA templates. This method not only enhances precision but also enables simultaneous execution of diverse edits, offering a more comprehensive toolkit for genetic manipulation.
The components of Prime Editing encompass a prime editing guide RNA (pegRNA) and a fusion protein comprising a Cas9 H840A nickase and a reverse transcriptase (Figure 1). The pegRNA serves a dual role, functioning as both a guide for identifying target sequences and a template for encoding desired genetic alterations. On the other hand, the fusion protein, featuring a modified Cas9 nickase and a reverse transcriptase, catalyzes the editing process with remarkable precision.
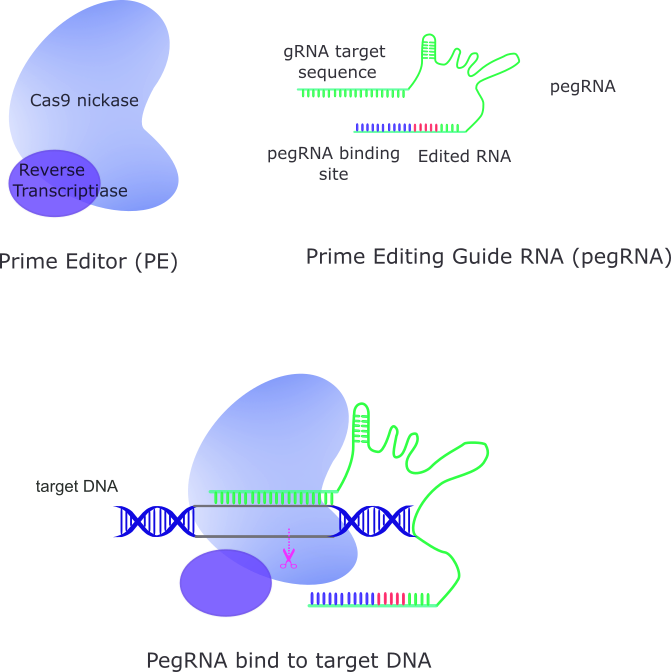
Figure 1. Components of Prime Editing and the Binding of pegRNA to Target DNA
Prime Editing operates through an intricate yet efficient mechanism. Initially, the pegRNA directs the prime editor protein to the target site, where the Cas9 nickase introduces a single-strand nick in the DNA molecule. Subsequently, the reverse transcriptase utilizes the pegRNA template to synthesize the desired genetic edits, effectively replacing the original DNA sequence. This process results in the formation of a heteroduplex DNA, consisting of one edited strand and one unedited strand, which undergoes resolution to favor incorporation of the edit into the unedited strand, thereby completing the editing process.
The advent of Prime Editing heralds several advantages over conventional gene-editing techniques. It exhibits reduced constraints concerning the location of protospacer adjacent motif (PAM) sequences, allowing for edits near or far from PAM sites. Moreover, Prime Editing offers unparalleled versatility and precision, facilitating a broader spectrum of base-to-base changes compared to traditional base editing methods. Additionally, it demonstrates superior efficiency and generates fewer byproducts than homology-directed repair (HDR), presenting a promising alternative for precise genetic modifications.
However, despite its transformative potential, it's imperative to acknowledge the limitations and challenges associated with Prime Editing. Optimization across different cell types and consideration of ethical implications remain critical aspects of its development and application.
Feel free to reach out at info@pixelbiosciences.com and at order@pixelbiosciences.com for inquires.

