Over the past few decades, advancements in cultivation methods and the rapid development of new and advancing technologies has been the driving force behind the “-omics era” and has greatly enhanced the rate of discovery for a myriad of microorganisms. However, applications of microarray and Next Generation Sequencing (NGS) technologies typically are very limited in determining the relative abundances of individual targets and visualization of cells, which are features of which fluorescence in situ hybridization (FISH) excels. For that reason, scientists have frequently turned to FISH as an effective methodology for driving microbiome research around the world.
Importance of microbiome study
Microbes are the ubiquitous, microscopic living creatures found in almost every different habitat on planet Earth. From the deepest sea trenches to your very own mucus membranes, microbes maintain an incredible range of biodiversity and persist in a wide array of harsh environmental conditions. Microbial study permits human beings to obtain a better understanding of diseases, ecological factors, and public health matters in order to gain an enhanced comprehension in the microscopic interactions around us.
The study of microbiomes, which constitutes an analysis of the behavior, interaction, and function of communities of microbes occupying a distinct environment, has rapidly expanded over the previous few decades. As a complex network of inter-special interaction, microbiome research gives us the clearest avenue to help understand a variety of things, such as developing concepts in human health and rising concerns about the wellbeing of our individual ecosystems at-large.
Specifically pointing to microbial populations and their impact on humans, studies on diverse landscape of microbiota in our gut has opened the door to an abundance of medical advancements. FISH has played a critical role in promoting the visualization of our internal ecosystems, allowing us to understand the more finite interactions our health has with the taxonomic diversity and spatial distribution of gut microbiota. By expanding our knowledge of host-microbial interactions in humans, the study of microbiomes proves critical in establishing further discoveries impacting public health.
Background on FISH and its application to microbiome study
In situ hybridization, as an early precursor to the fluorescent methodology discussed in this post, was developed as a unique staining approach by which a reporter-labeled nucleotide probe was bound to a complimentary nucleic acid sequence target. With years of development following its initial formulation, in situ hybridization quickly began to advance, adopting a method for fluorescence detection and being widely implemented for microbiological study by the late 1980s.
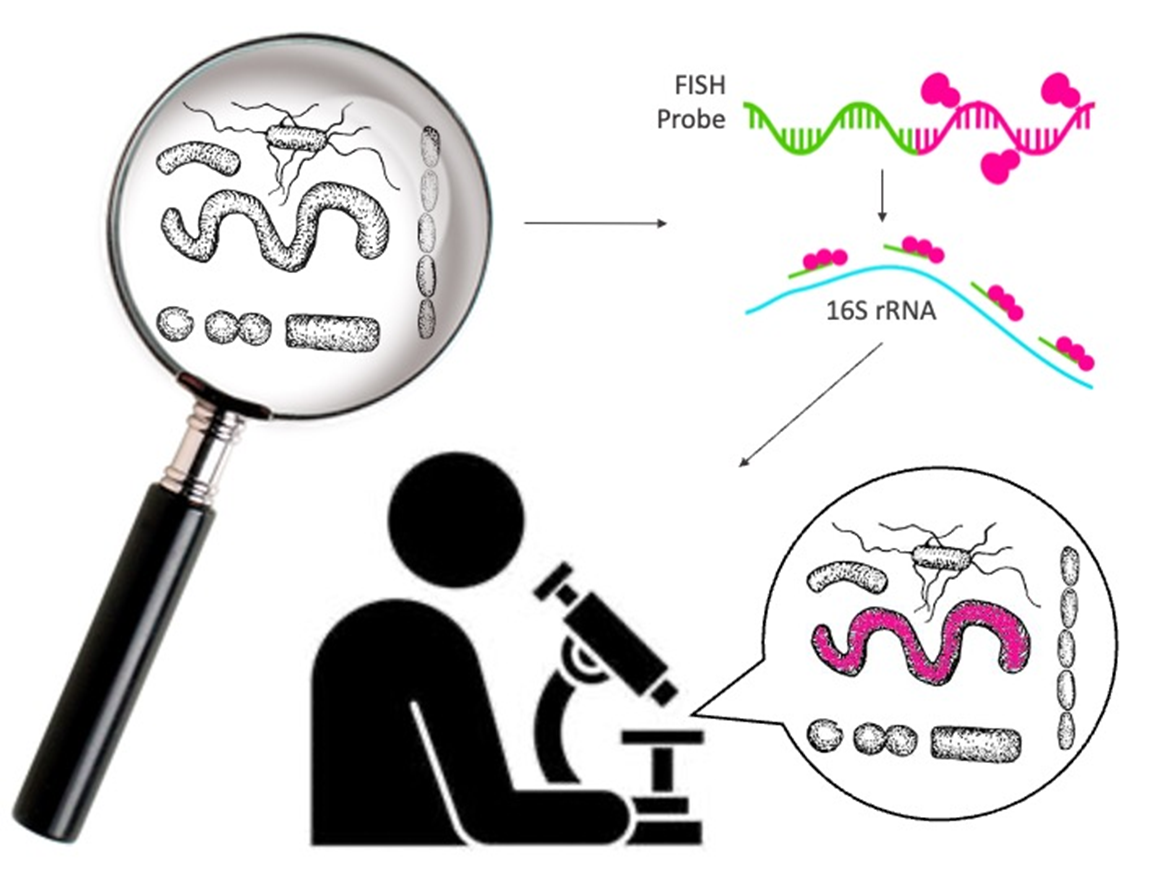
In the previous few decades, FISH has a stood as a commonly utilized and formidable tool for the analysis of specific groups of microorganisms. The modern practice utilizes labelled DNA or RNA probes to target ribosomal RNA (rRNA) of defined taxonomic groups. Commonly, 16S rRNA genes are often targeted in microbiome studies thanks to the abundance of both conserved and taxon-specific regions. The fluorescent dye-tagged probe is bound to a complimentary sequence present in the cell, and the subsequent fluorescence can be visualized as a determining factor of the presence and relative abundance of a target taxon in a particular microbiome environment.
Benefits of FISH analysis to microbiome study
When dealing with microbiomes, it is critical to remember that you aren’t simply dealing with a single species, but rather an entire ecosystem of diverse and exotic microbes, all existing in different volumes within a specified setting. As such, utilizing a culture-dependent analysis method, such as FISH, has some specific advantages.
FISH, being sensitive and semi-quantitative, permits for a higher level of detection for low-abundant organisms in an environment, permitting for a more complete observation of the biodiversity of a particular microbiome. It additionally excels in the detection of extracellular organisms located within host cells, especially when attempting to target sparser species that might be difficult to grow in media. The ability to have a sensitive analysis methodology when observing complex inter-special networks is critical in getting a comprehensive understanding of the environment being analyzed.
In addition to our general understanding of FISH and its functionality, researchers across the globe have been able to augment the typical FISH methodology in order to obtain more specific or sensitive data pointed at different matrix targets.
For example, single molecular fluorescence in situ hybridization (smFISH) has been widely used in research across a variety of fields. It functions by permitting multiple probes, all conjugated to the same fluorescent dye, to target an individual molecule of RNA. Through binding of multiple fluorescent probes to the singular RNA molecule, the signal is systematically increased as more probes are complimentarily bound, effectively improving the signal-to-noise ratio that can be detrimentally impacted by non-specific binding.
As of late, smFISH applications have been used to study the impact of the gut microbiome on Alzheimer’s disease progression, in addition to a study on the impact of an innate viral immune response on microbiome health.
Comparison of the use of FISH and other analytical methods
When it comes to the taxonomic profiling of microbiomes, scientists have utilized a multitude of methods. Nucleic acid-based microbiome analysis, such as that conducted by microarrays and Next Generation Sequencing, are generally utilized in the field. While both methodologies do have their advantages, both come at a significantly higher expense when compared to culture-focused techniques like FISH. This permits the utilization of FISH for microbiome study to be more advantageous in situation of resource-limiting, whereas both microarrays and Next Generation Sequencing require heavy financial commitments in order to utilize.
Despite having a higher potential throughput than culture-dependent assays like FISH, Next Generation Sequencing is limited in terms of quantitative analysis, failing to be able to accurately determine the absolute abundance of a taxon in an environment.
However, researchers and laboratories also frequently combine multiple applications in order to obtain a myriad of information regarding nucleic acid targets. By combining both NGS and in situ hybridization methods, researchers have been able to sequence swarms of microbes, before turning to in situ hybridization as a means of furthering their understanding on the morphological context and association of those microbes within a target environment.
Between microbial study and a wide range of other applications, FISH is an inexpensive and reliable method for the analysis of target nucleic acid sequences. Its role in microbiome study stands with its sensitivity and constant progression since its inception over 40 years ago, making it a versatile methodology in multiple fields of study.
When it comes to microbiome study with FISH, probe design tends to be the parameter that has a stronger effect on the sensitivity and specificity of your methodology. Reach out to us an let us help you with your upcoming FISH project: order@pixelbiosciences.com

