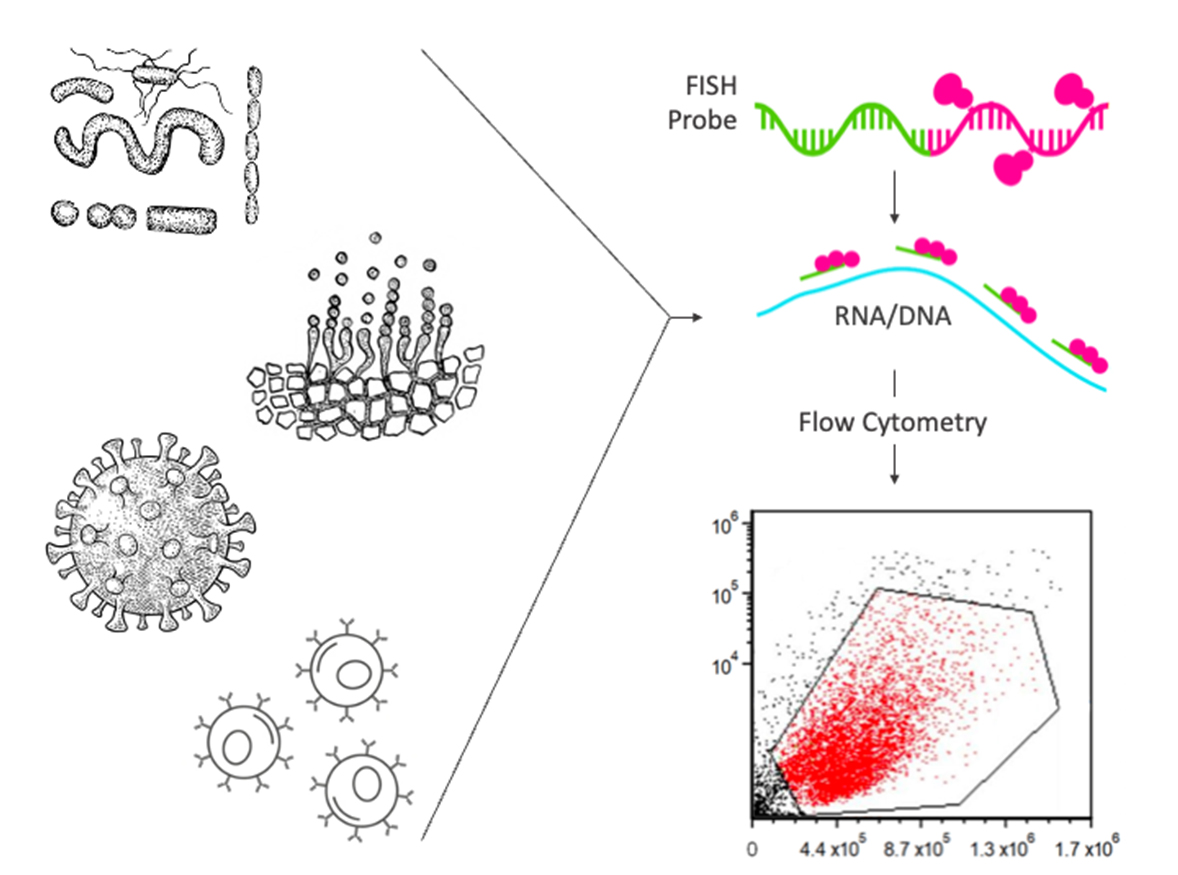
Several FISH-,qPCR- or RNA sequencing-based methods can measureRNA expression at the single-cell level. One technique that is suitablefor high-throughput analysis is flow cytometry-based fluorescence in situ hybridization, or Flow-FISH.
It’s design process is relatively straightforward, making Flow-FISH a valuable tool where no good fluorescently-labeled antibodies are avail-able for the target of interest (e.g., difficult to stain cytokines), when no protein product is formed (e.g., noncoding RNAs), or when studying model organisms for which the antibody has not yet been developed. Moreover,in comparison to antibodies that can suffer from cross reac- tivity, limited affinity, and poor signal, FISH probes work in highly specific manner due to nucleotide base pairing. Additionally,Flow-FISH assays can also be combined with fluorescently labeled antibodies allowing for the measurement of mRNA and protein of the same gene at.
Flow-FISH applications
Bacteria
Flow-FISH probes directed at specific bacterial sequences allow for identification of individual bacterial species preventing the need for,e.g.,extensive culturing. Potential applications include cell sorting of live bacteria based on gene expression followed by downstream analyses of sorted cells, such as the importance of gene(s) of interest on, e.g., growth patterns, bacterial virulence, and/or antibiotic resistance. Similarly,Flow-FISH may fa-cilitate identification of bacterial infections or bacterial food contamination.
Fungi
Similar to bacterial Flow-FISH assays, also in fungi, Flow-FISH offers the advantage of detecting fungal strains, assess food contamination, investigate infection models, and to study antimycotic resistance.
Other applications where Flow-FISH could be used to study fungal species include gene transcription/single-cell gene expression based on stimuli or growth conditions under diverse conditions.
Viruses
While Flow-FISH has been employed to study both bacteria and fungi, it has mostly been used as a tool to study a variety of viruses, including both human pathogens,, and other organisms like mice, cattle or simians. These applications include the detection of infected cells, viral strain identification, and studying viral biology.
Due to its single-cell approach, Flow-FISH assays provide more information compared to conventional diagnostic tests. Furthermore, Flow-FISH has also been to study pathogens responsible for emerging diseases, such as dengue virus,SARS-CoV-2,and Zika virus.
Cells
To date, Flow-FISH has been used in T cells for measuring telomere length and studying T cell functions.Therefore, Flow-FISH may be of par-ticular interest in the study of effector molecules that are notoriously difficult to detect with classical antibody-mediated or secretion essays, like cytokines and chemokines.
Additionally, Flow-FISH is also suitable for detection of transcription factors. Furthermore,in combination with specific surface markers, Flow-FISH could help identify the T cell population and potentially help unravel the source of effector molecules from different immune cells during the course of infection.
Single-Molecule Flow-FISH with Multiple Probes
Flow-FISH assay that is often used is single-molecule Flow-FISH with multiple probes. Typically, these probes are 20-nucleotide-long single-strand oligos that are designed to specifically anneal to a single DNA/RNA molecule. The probe design is fairly simple and we developed a pipeline to design and filter for oligos with high hybridization efficiency, a better sig-nal-to-noise ratio and higher contrast. We apply a novel cost-effective, enzymatic fluorophore labeling assay and in principle, our method also allows for multiplex barcoding of the probe, enabling to detect 7 genes of interest in one reaction, not only on mRNA level, but also DNA.
Flow-FISH with multiple smFISH probes is inexpensive, fast, and easy to implement but identifying the background staining is critical for the setup of the assay. Validation of the smFISH signal can be performed with unlabeled sequence-identical oligos which compete with the fluo- rescently labeled RNA probes for binding. Alternatively,background fluorescence levels can also be determined by using non relevant probes that are labeled with the same fluorochrome as the probe set of interest. In fact, the number of simultaneous RNA measurements is only limited by the detection channels on the flow cytometer.
References:
Pereira et aL.,When FLOW-FISH met FACS: combining multiparametric, dynamic approaches for microbial single-cell research in the total environment.Sci-ence of the Total Environment.2022
Freen-van Heeren, Flow-FISH as a Tool for Studying Bacteria,Fungi and Viruses.BioTech.2021
Freen-van Heeren et al., Measuring Tcell Responses by Flow Cytometry-Based Fluorescence In Situ Hybridization.Critical Reviews in Immunology.2018

